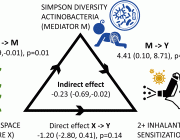
Synthetic Symphony: Scientists Evoke Artificial DNA Transcription with Natural Enzyme Expertise
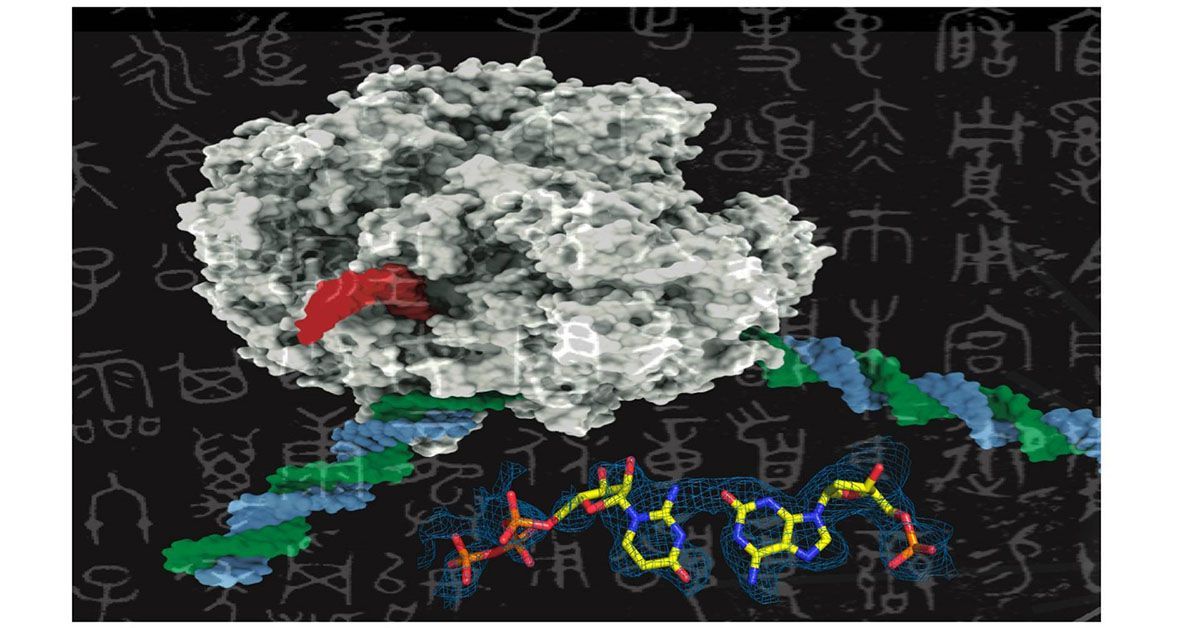
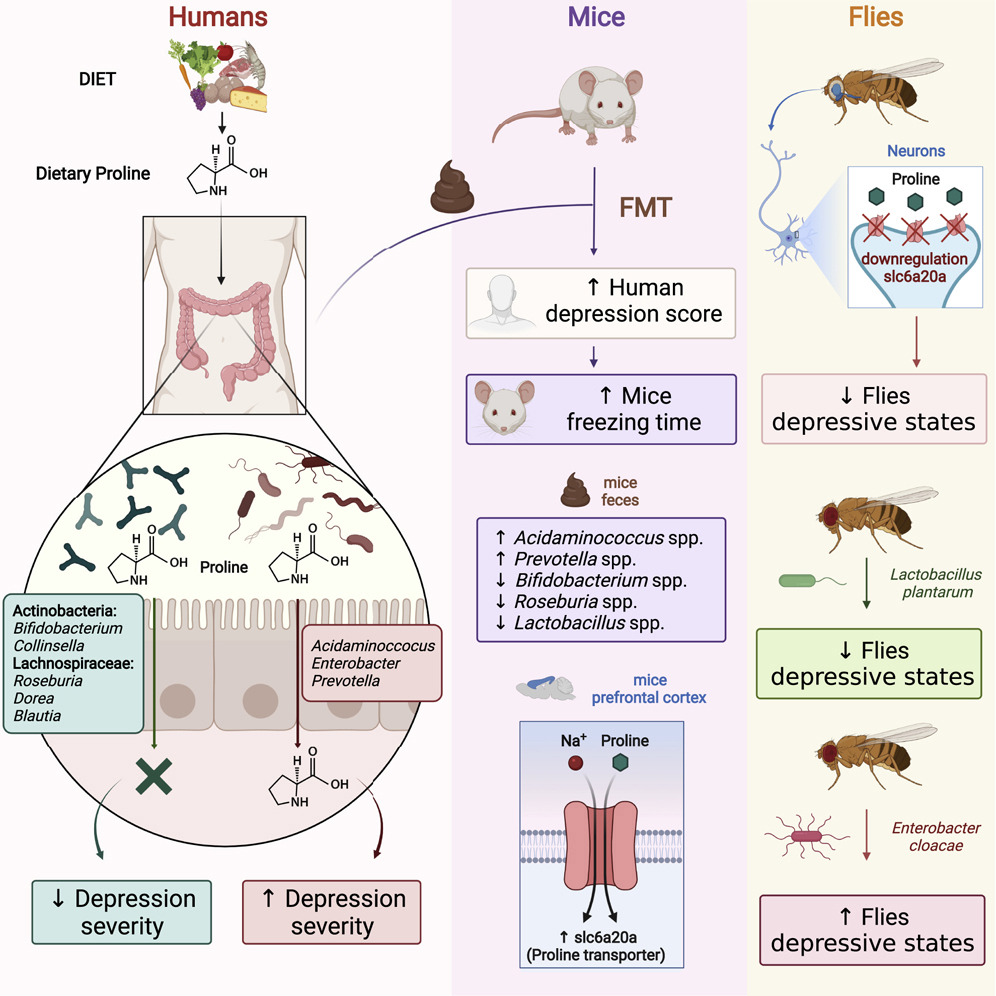
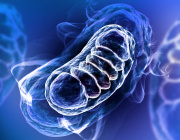
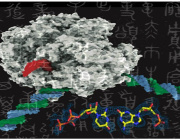
Like adding new letters to an existing language’s alphabet to expand its vocabulary, adding new synthetic nucleotides to the genetic alphabet could expand the possibilities of synthetic biology. This image shows a rendering of RNA polymerase (center) and a synthetic nucleotide (lower right). Photo credits: UC San Diego Health Sciences
A groundbreaking study recently emerged from a collaborative effort by researchers at the University of California San Diego, Salk Institute for Biological Sciences, and Shuichi Hoshika at the Foundation for Applied Molecular Evolution. The research delves into the intriguing domain of Artificially Expanded Genetic Information Systems (AEGIS), providing crucial insights into the interplay of these systems with cellular RNA polymerases (RNAPs) within the context of E. coli.
AEGIS introduces independently replicable unnatural nucleotide pairs alongside the naturally occurring G:C and A:T/U pairs found in native DNA. The study meticulously explains the mechanisms through which E. coli RNAP selectively recognizes these unnatural nucleobases within a six-letter expanded genetic system. Utilizing high-resolution cryo-EM structures, the researchers meticulously capture three RNAP elongation complexes containing template-substrate Unnatural Base Pairs (UBPs), revealing fundamental principles governing the recognition of both AEGIS and natural base pairs.
In these structural depictions, RNAPs are portrayed in an active state, poised to execute the crucial chemistry step. Notably, at this pivotal juncture, the unnatural base pair adopts a Watson-Crick geometry, and the trigger loop assumes an active conformation. This groundbreaking revelation signifies that the mechanistic principles dictating the recognition and incorporation of natural base pairs are equally applicable to AEGIS unnatural base pairs, thereby validating the underlying design philosophy of AEGIS.
The researchers contribute additional structural evidence, affirming a longstanding hypothesis that pair mismatch during transcription occurs via tautomerization. This nuanced insight emphasizes the significance of Watson-Crick complementarity in shaping the design principles governing AEGIS base pair recognition.
This groundbreaking work transcends the boundaries of our understanding of genetic information systems, affirming the resilience of Watson-Crick complementarity in recognizing both artificial and natural base pairs. These findings carry profound implications for the future design and engineering of genetic information, heralding a new era in synthetic biology.
Learn more about this fascinating study.
International Society of Microbiota
LinkedIn | Facebook